- [11-05]freemarker模板中long类型问题_qq_31459039的博客
- [07-27]怎么解决WB实验显影方面的问题 ?_答魔科研
- [06-03]小鼠巨噬细胞M1、M2分型表面标记 基础问题 流式中文网
- [11-05]marker是什么意思_marker蚂蚁淘进口试剂平行进口
- [12-29]激光共聚焦科技高清完整正版视频在线观看优酷
- [06-15]western marker跑不开原因求助 蛋白质和糖学技术讨论版论坛
- [07-31]...蛋白转到膜上了,但是marker看不到,为什么marker没转上呢?
- [05-30]又出新问题……WB转膜后marker不见了…… 蛋白质和糖学技术讨论...
- [04-11]【干货分享】WB实验问题总结与处理方案
Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer | Molecular Cancer | Full Text
Abstract
LongnoncodingRNAHOTTIPplaysimportantrolesinthegenerationandprogressionofhumancancers.Exosomesparticipateincellularcommunicationbytransmittingmolecularsbetweencellsandareregardedassuitablecandidatesfornon-invasivediagnosis.However,theexistenceofHOTTIPinthecirculatingexosomesandthepotentialrolesofexosomalHOTTIPingastriccancer(GC)waspoorlyunderstood.ThisstudyaimsatinvestigatingtheclinicalrolesofexosomalHOTTIPinGC.SerumexosomalHOTTIPfrom246subjects(126GCpatientsand120healthypeople)weredetectedbyreversetranscriptionreal-timequantitativepolymerasechainreaction(RT-qPCR).OurresultsshowedthatexpressionlevelsofexosomalHOTTIPweretypicallyupregulatedinGCthaninnormalcontrol(P鈥?lt;鈥?.001).Anditsexpressionlevelsweresignificantlycorrelatedwithinvasiondepth(P鈥?鈥?.0298)andTNMstage(P鈥?lt;鈥?.001).TheAUCforexosomalHOTTIPwas0.827,whichdemonstratedahigherdiagnosticcapABIlitythanCEA,CA19鈥?andCA72鈥?(AUC鈥?鈥?.653,0.685and0.639,respectively)(P鈥?lt;鈥?.001).TheKaplan鈥揗eieranalysisshowedacorrelationbetweenincreasedexosomalHOTTIPlevelsandpooroverallsurvival(OS)(logrankP鈥?lt;鈥?.001).AndunivariateandmultivariateCOXanalysisrevealedexosomalHOTTIPoverexpressionwasanindependentprognosticfactorinGCpatients(P鈥?鈥?.027).ThesefindingsdemonstratedthatexosomalHOTTIPmaybeapotentialbioMarkerforGCindiagnosisandprognosis.
Gastriccancer(GC)isahugeburdenworldwidewithhighmorbidityandhighmortality,especiallyinEasternAsia[1].Despitemanyadvancesinthediagnosisandtreatment,theprognosisofGCpatientsremainspoor,withover70%patientsultimatelydiedfromthisdisease[2].EarlydiagnosisandearlytreatmentisconducivetoimprovingprognosisofGC.ThegoldstandardforGC,endoscopyfollowedbypathologicalexamination,isinvasiveanduncomfortable[3].Bodyfluids-basedtestingisconsideredtobeasatisfactorymethodwithoutinvasion,butcurrentserumtumorbiomarkersusedforGCdiagnosis,likeCEA,CA19鈥?andCA72鈥?,havealowpositiverate[4].Theserealitieshighlightthenecessitytodiscoverothernovelbiomarkerswithhighspecificityandhighsensitivity.
Longnon-codingRNAs(lncRNAs)areagroupofnon-codingRNAswithalengthlongerthan200nucleotides[5].EvidencesshowdisorderlyregulationoflncRNAstakespartinhumandisease[6].TheHOXAtranscriptatthedistaltip(HOTTIP),whichtranscribedfromthe5鈥?tipofHOXAcluster,hasgainedgrowingattentionamongcancer-relatedlncRNAs[7].HOTTIPprimarilytargetsWDR5/MLLcomplexesacrossHOXAviadirectlybindingtheadaptorproteinWDR5,leADInghistoneH3lysine4trimethylationandgenetranscriptionofseveral5鈥?HOXAgenesandreportshaveshowedHOTTIPcanpromotecellproliferationthroughp21inhibitionormiRNAsilencing[8,9,10].Yeetal.[11]foundthatHOTTIPwassignificantlyoverexpressedinGCcelllinesanddownregulationofHOTTIPwouldinhibitcellproliferation,promotecellapoptosis,anddegradecellmigrationandinvasion.StudiesofGCtissuesrevealedthatupregulatedexpressionsofHOTTIPwasassociatedtopoordifferentiation,positivelymphnodesmetastasisandadvancedclinicalstage[12].However,nostudyofHOTTIPinGCserumhasbeenreported.
Exosomesareregardedasanentirelynewmodeofcell-to-cellcommunicationwithasizeof50-150聽nmindiameter,whicharesecretedbymanycellsandcanreflectthecharacteristicsoftheirparentcells[13,14].Exosomescontainvariousandnumerouscargos,includingenzymes,proteins,lipidsandRNAs,andpresentinperhapsallBIOLOGicalfluids,thustheyarebelievedtoparticipateinmultiplepathologicalconditions,includingcancer[15,16].FurThermore,exosomaldoublelayermembraneisanidealshelterforprotectingthecargoesfrombeingdegradedandthestabilityandlongevityoftheRNAmoleculesareidealfornon-invasivediagnosisoftumor[17].Inpreviousstudy,ourlabhadprovedtheexistenceandstabilityofexosomallncRNAinserumanddetailedtheclinicalsignificanceofexosomallncRNACRNDE-hforCRC[17].Inviewoftheseobservations,wesupposedthatGCcellsareabletogenerateandreleaseexosomesandtheexaminationofexosomalHOTTIPmayprovideanoninvasivemethodforGCdetection,patientssubpopulation,prognosispredictionandevenallowbetterunderstandingofetiopathology.Inthisstudy,wesystematicallyinvestigatedtheexpressionofexosomalHOTTIPinGCserumandtappedinformationmaybeprovidedabouttheGCstatus.ThenwefurthertestedthefeasibilitythatHOTTIPasapotentialnovelmarkerforGCdiagnosisandprognosisandcomparedthediagnosticcapabilityofHOTTIPwithtraditionalbiomarkers,includingCEA,CA19鈥?andCA72鈥?.
ResultsanddiscussionExpressionofexosomalHOTTIPanditsrelationshipwithclinicopathologicalfesturesinGCpatientsInthisstudy,wefoundthatexosomalHOTTIPcouldbedetectedinculturemediumofGCSGC7901cellline,andthelevelswereincreasedwiththeincubationtimeextended(Additional聽file聽1:FigureS1),suggestingexosomalHOTTIPwasdirectlyreleasedfromGCcells.
Then,weenrolled126GCpatientsand120normalhealthysubjectsbetweenJanuary,2011andMay,2012atQiluHospital,ShandongUniversity,China.Serumsampleswerecollectedbeforesubjectshadreceivedanysurgery,chemotherapyorradiationtherapy.Andthen,theyweresub-packedintoEppendorftubeswithoutRnase,Dnaseorpyrogenafterbeingcentrifugedat11000聽rpmfor10聽minandimmediatelyfrozenandstoredat鈭掆€?0聽掳Cuntilanalysis.LevelsofexosomalHOTTIPinthese246serumsamplesweredetectedbyRT-qPCRusingGAPDHandUBCasinternalcontrol.WefoundthatexosomalHOTTIPexpressionlevelswereevidentlyupregulatedinGCpatients(P聽鈥?.001)(Additional聽file聽2:FigureS2).ToobservethestabilityofexosomalHOTTIPinserum,wemeasuredtheirexpressionlevelsaftertreatedwithprolongedexposuretoroomtemperatureormutiplefreeze-thawcyclesandourresultsdemonstratedthatallthesetreatmenthadnoinfluenceontheirexpression(Additional聽file聽3:FigureS3).
WealsoanalyzedtheassociationbetweenexosomalHOTTIPexpressionlevelsandGCpatients鈥?clinicopathologicalfeatures.WefoundexosomalHOTTIPexpressionlevelswerepositivelyassociatedwithinvasiondepth(P鈥?鈥?.0298)andTNMstage(P鈥?lt;鈥?.001)ofGC.Conversely,therewasnosignificantcorrelationofexosomalHOTTIPexpressionwithotherclinicalfeatures,suchasage,gender,sizeoftumor,pathologicaldifferentionandlymphnodesmetastasis(allP鈥?gt;鈥?.05)(Table聽1).
Table1CorrelationbetweenexosomalHOTTIPandclinicopathologicalparametersofgastriccancer(n鈥?鈥?26)FullsizetableDiagnosticvalueofexosomalHOTTIPforGCpatientsCEA,CA19鈥?andCA72鈥?arewidelyusedinscreeningandauxiliarydiagnosinggastrointestinalmalignancies[4].WemeasuredtheirexpressionlevelsinGCserumbyelectRochemiluminescencemethodonCobasE601(RocheDiagnosticsGmbH,Germany)andconstructedROCcurvestoevaluateandcomparedthediagnosticcapacityofserumbiomarkers.Theareaunderthecurve(AUC)forexosomalHOTTIPwas0.827(Fig.聽1a),whichsignificantlyhigherthantheAUCsforCEA,CA19鈥?,CA72鈥?with0.653,0.685and0.639,respectively(P鈥?lt;鈥?.001;Fig.聽1b,candd).WecalculatedthecombinativediagnosticvalueofCEA,CA19鈥?andCA72鈥?,theAUCwas0.723(Fig.聽1e).AndtheresultsalsoshowedtheAUCforexosomalHOTTIPwassignificantlylargerthanthatfortheseunion(P鈥?lt;鈥?.001),indicatingthatexosomalHOTTIPwassuperiortotheminGCdiagnosis.WefurtherevaluatedthecombinativediagnosticvalueofCEA,CA19鈥?,CA72鈥?andexosomalHOTTIPforGC.TheAUCforthiscombinationwas0.870(Fig.聽1f).Thecombinativediagnosticcapabilitywasbetterthanoneofthesemarkersalone.Attheoptimalcut-offvalue(1.720),thesensitivityandspecificityofexosomalHOTTIPwere69.8and85.0%,respectively.Whenwesetthespecificityas95%,exosomalHOTTIPhadthehighestsensitivity(Additional聽file聽4:TableS1).TheseresultsindicatedthatexosomalHOTTIPmightbeanappropriatediagnosticmarkerforGC.
Fig.1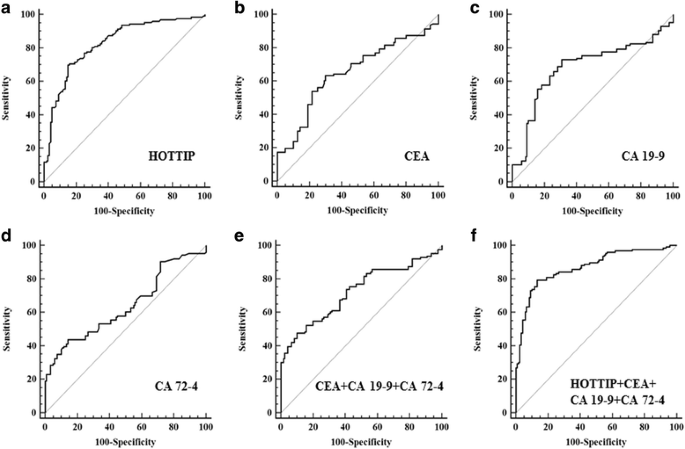
TheROCcurvesofbiomarkers.aTheROCcurveofexosomalHOTTIP.Theareaunderthecurvewas0.827.bTheROCcurveofCEA.Theareaunderthecurvewas0.653.cTheROCcurveofCA19鈥?.Theareaunderthecurvewas0.685.dTheROCcurveofCA72鈥?.Theareaunderthecurvewas0.639.eTheROCcurveofcombineCEA,CA19鈥?andCA72鈥?.Theareaunderthecurvewas0.723.fTheROCcurveofcombineCEA,CA19鈥?,CA72鈥?andexosomalHOTTIP.Theareaunderthecurvewas0.870.P鈥?lt;鈥?.001
FullsizeimagePrognosticvalueofexosomalHOTTIPforGCpatientsEmergingevidenceshowedthatHOTTIPwasupregulatedandcorrelatedtopoorprognosisinpatientswithcancers[18].Inourstudy,Kaplan鈥揗eiermethodwasperformedtoanalyzetherelationshipsbetweenCEA,CA19鈥?,CA72鈥?andexosomalHOTTIPexpressionandoverallsurvivalrateofpatientswithGC.TheresultsdemonstratedthattherewasnosignificantrelationshipbetweenthesetraditionaltumormarkersandOS(allP聽鈥?.05;Fig.聽2a,bandc).However,patientswithhighexosomalHOTTIPexpressionhadatendencyofshorter5-yearoverallsurvival(P鈥?lt;鈥?.001;Fig.2d).
Fig.2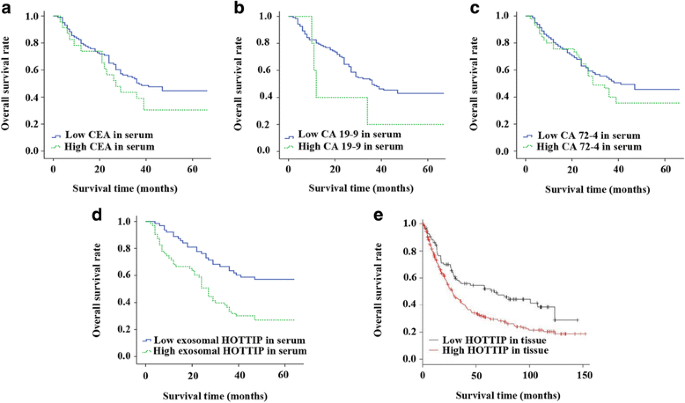
Associationbetweentumormarkersandoverallsurvivalingastriccancer.Kaplan鈥揗eiercurvesofgastriccancerpatientsaccordingtoserumlevelsofaCEA,bCA19鈥?,cCA72鈥?anddexosomalHOTTIP,andelevelsofHOTTIPintissue
FullsizeimageTheonlinedatabaseK-Mplotter(www.kmplot.com),basingonthedetectiondatafromGCtissues,wasusedtovalidatepatientsurvivalrate.Onethousandandsixty-fivetissuesamplesofGCwereincludedinK-Mplotterdatabaseandthepatients鈥?informationswereassembledfromGEO(Affymetrixmicroarraysonly),EGAandTCGA.Themeanfollow-uptimeofthesepatientswas33聽months[19].WecomputedprognosticvalueofHOTTIPbyusingtheAffymetrixprobeatameanprobeof1564069_at,244553_atand1564070_s_atandchoosingfivedatabases(GSE14210,GSE15459,GSE22377,GSE29272,GSE51105).Finally,348patients鈥?datawereemployed.Thepatientswereclassifiedintotwogroupsonthebasisofvariousquantileexpressionsoftheproposedbiomarker.Meanwhile,thehazardratioandlogrankPvaluewerecalculated.TheresultofK-MplottersurvivalanalysisdemonstratedthatexpressionlevelsofHOTTIPwereincreasedabovetheoptimalcutoffpointin71.8%ofpatientswithGCandthesepatientshadapooroverallsurvival,withthepooledHRof1.63(95%CI鈥?鈥?.19鈥?.23,P鈥?鈥?.0022)(Fig.聽2e),whichsupportedourconclusion.
Coxproportionalhazardsmodelwerecalculatedtoidentifyindependentprognosticfactors.UnivariateanalysisindicatedthatadvancedTNMstage(HR鈥?鈥?.000,95%CI鈥?鈥?.225鈥?.264,P鈥?鈥?.006)andhighexosomalHOTTIPlevels(HR鈥?鈥?.352,95%CI鈥?鈥?.460鈥?.791,P鈥?lt;鈥?.001)werepoorprognosticfactors(Additionalfile3:FigureS3).ThenfeatureswithavalueofP鈥?lt;鈥?.05inunivariateanalysiswereputintomultivariateanalysis.TheanalysisshowedthathighexosomalHOTTIPlevelswereindependentfactorsassociatedwithpoorOSofGC(HR鈥?鈥?.037,95%CI鈥?鈥?.085鈥?.823,P鈥?鈥?.027)(Additional聽file聽5:TableS2).
ConclusionsOurstudyrevealesthepresenceofHOTTIPinexosomesisolatedfromserumsamplesofGCpatients.WefindthatexosomalHOTTIPisupregulatedintheserumofGCpatientsanditmaybeabetterbiomarkerforGCindiagnosisthanCEA,CA19鈥?andCA72鈥?.Furthermore,exosomalHOTTIPexpressionlevelsisidentifiedasanindependentfactorforpoorprognosisinGCpatients.Inconclusion,exosomallongnoncodingRNAHOTTIPmaybeapotentialbiomarkerforgastriccancerindiagnosisandprognosis.
AbbreviationsCA19鈥?:Carbohydrateantigen19鈥?
CA72鈥?:Cancerantigen72鈥?
CEA:Carcinoembryonicantigen
GAPDH:Glyceraldehyde-3-phosphatedehydrogenase
Gastriccancer
HOTTIP:TheHOXAtranscriptatthedistaltip
UBC:UbiquitinC
References1.GlobalBurdenofDiseaseCancerC,FitzmauriceC,DickerD,PainA,HamavidH,Moradi-LakehM,MFMI,AllenC,HansenG,WoodbrookR,etal.TheGlobalBurdenofCancer2013.JAMAOncol.2015;1:505鈥?7.
ArticleGoogleScholar2.AllemaniC,WeirHK,CarreiraH,HarewoodR,SpikaD,WangX-S,BannonF,AhnJV,JohnsonCJ,BonaventureA,etal.Globalsurveillanceofcancersurvival1995鈥?009:analysisofindividualdatafor25鈥?76鈥?87patientsfrom279population-basedregistriesin67countries(CONCORD-2).Lancet.2015;385:977鈥?010.
ArticlePubMedGoogleScholar3.ParkJY,vonKarsaL,HerreroR.Preventionstrategiesforgastriccancer:aglobalperspective.ClinEndosc.2014;47:478鈥?9.
ArticlePubMedPubMedCentralGoogleScholar4.ShimadaH,NoieT,OhashiM,ObaK,TakahashiY.Clinicalsignificanceofserumtumormarkersforgastriccancer:asystematicreviewofliteraturebythetaskforceoftheJapanesegastriccancerassociation.GastricCancer.2014;17:26鈥?3.
CASArticlePubMedGoogleScholar5.BonasioR,ShiekhattarR.RegulationoftranscriptionbylongnoncodingRNAs.AnnuRevGenet.2014;48:433鈥?5.
CASArticlePubMedPubMedCentralGoogleScholar6.DenizE,ErmanB.LongnoncodingRNA(lincRNA),anewparadigmingeneexpressioncontrol.FunctIntegrGenomics.2017;17:135鈥?3.
CASArticlePubMedGoogleScholar7.LianY,CaiZ,GongH,XueS,WuD,WangK.HOTTIP:acriticaloncogeniclongnon-codingRNAinhumancancers.MolBioSyst.2016;12:3247鈥?3.
CASArticlePubMedGoogleScholar8.WangKC,YangYW,LiuB,SanyalA,Corces-ZimmermanR,ChenY,LajoieBR,ProtacioA,FlynnRA,GuptaRA,etal.AlongnoncodingRNAmaintainsactivechromatintocoordinatehomeoticgeneexpression.Nature.2011;472:120鈥?.
CASArticlePubMedPubMedCentralGoogleScholar9.LianY,DingJ,ZhangZ,ShiY,ZhuY,LiJ,PengP,WangJ,FanY,DeW,WangK.ThelongnoncodingRNAHOXAtranscriptatthedistaltippromotescolorectalcancergrowthpartiallyviasilencingofp21expression.TumourBiol.2016;37:7431鈥?0.
CASArticlePubMedGoogleScholar10.GeY,YanX,JinY,YangX,YuX,ZhouL,HanS,YuanQ,YangM.MiRNA-192[corrected]andmiRNA-204directlysuppresslncRNAHOTTIPandinterruptGLS1-mediatedGlutaminolysisinhepatocellularcarcinoma.PLoSGenet.2015;11:e1005726.
ArticlePubMedPubMedCentralGoogleScholar11.YeH,LiuK,QianK.OverexpressionoflongnoncodingRNAHOTTIPpromotestumorinvasionandpredictspoorprognosisingastriccancer.OncoTargetsTher.2016;9:2081鈥?.
PubMedPubMedCentralGoogleScholar12.ChangS,LiuJ,GuoS,HeS,QiuG,LuJ,WangJ,FanL,ZhaoW,CheX.HOTTIPandHOXA13areoncogenesassociatedwithgastriccancerprogression.OncolRep.2016;35:3577鈥?5.
CASArticlePubMedGoogleScholar13.MeloSA,LueckeLB,KahlertC,FernandezAF,GammonST,KayeJ,LeBleuVS,MittendorfEA,WeitzJ,RahbariN,etal.Glypican1identifiescancerexosomesandfacilitatesearlydetectionofcancer.Nature.2015;523:177鈥?2.
CASArticlePubMedPubMedCentralGoogleScholar14.KahlertC,MeloSA,ProtopopovA,TangJ,SethS,KochM,ZhangJ,WeitzJ,ChinL,FutrealA,KalluriR.Identificationofdouble-strandedgenomicDNAspanningallchromosomeswithmutatedKRASandp53DNAintheserumexosomesofpatientswithpancreaticcancer.JBiolChem.2014;289:3869鈥?5.
CASArticlePubMedPubMedCentralGoogleScholar15.PantS,HiltonH,BurczynskiME.Themultifacetedexosome:biogenesis,roleinnormalandaberrantcellularfunction,andfrontiersforpharmacologicalandbiomarkeropportunities.BiochemPharmacol.2012;83:1484鈥?4.
CASArticlePubMedGoogleScholar16.TkachM,TheryC.Communicationbyextracellularvesicles:whereweareandwhereweneedtogo.Cell.2016;164:1226鈥?2.
CASArticlePubMedGoogleScholar17.LiuT,ZhangX,GaoS,JingF,YangY,DuL.ExosomallongnoncodingRNACRNDE-hasanovelserum-basedbiomarkerfordiagnosisandprognosisofcolorectalcancer.Oncotarget.2016;7:85551鈥?3.
PubMedPubMedCentralGoogleScholar18.YangY,QianJ,XiangY,ChenY,QuJ.TheprognosticvalueoflongnoncodingRNAHOTTIPonclinicaloutcomesinbreastcancer.Oncotarget.2017;8:6833鈥?4.
PubMedGoogleScholar19.Sz谩szAM,L谩nczkyA,Nagy脕,F枚rsterS,HarkK,GreenJE,BoussioutasA,BusuttilR,Szab贸A,Bal谩zsG.Cross-validationofsurvivalassociatedbiomarkersingastriccancerusingtranscriptomicdataof1,065patients.Oncotarget.2016;7:49322鈥?3.
ArticlePubMedPubMedCentralGoogleScholarDownloadreferences
FundingThisstudywassupportedbyShandongKeyResearchandDevelopmentProgram(2016GSF201122,2016CYJS01A02),NaturalScienceFoundationofShandongProvince(ZR2017MH044),FundamentalResearchFundsofShandongUniversity(2014QLKY03),NationalNaturalScienceFoundationofChina(81601846),andTaishanScholarFoundation.
AvailabilityofdataandmaterialsTheauthorsdeclarethatdatasetssupportingtheconclusionsofthisstudyareavailablewithinthemanuscriptanditssupplementaryinformationfiles.
AuthorinformationAuthornotesAffiliationsDepartmentofClinicalLaboratory,QiluHospital,ShandongUniversity,107WenhuaXiRoad,Jinan,Shandong,250012,ChinaRuiZhao,聽XinZhang,聽YongmeiYang,聽XinZheng,聽XiaohuiLi,聽YingjieLiu聽聽YiZhangDepartmentofClinicalLaboratory,ShandongProvincialThirdHospital,Jinan,Shandong,250012,ChinaYanliZhangAuthorsRuiZhaoViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarYanliZhangViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarXinZhangViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarYongmeiYangViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarXinZhengViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarXiaohuiLiViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarYingjieLiuViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarYiZhangViewauthorpublicationsYoucanalsosearchforthisauthorinPubMedGoogleScholarContributionsXZandYZconductedthestudydesign;RZ,YZandYYperformedexperimentsandanalyzedthedata.YZ,XZ,XZ,XLandYLcollectedthesamples;RZ,YZandYYdraftedthemanuscript.Alltheauthorsreviewedandapprovedthefinalmanuscript.
CorrespondingauthorCorrespondencetoYiZhang.
EthicsdeclarationsEthicsapprovalandconsenttoparticipateThisstudywasreviewedandapprovedbytheEthicsCommitteeofQiluHospitalofShandongUniversity,andallpatientsprovidedwritteninformedconsent.
Publisher鈥檚Note
SpringerNatureremainsneutralwithregardtojurisdictionalclaimsinpublishedmapsandinstitutionalaffiliations.
Additionalfile
================ 蚂蚁淘在线 ================
免责声明:本文仅代表作者个人观点,与本网无关。其创作性以及文中陈述文字和内容未经本站证实,对本文以及其中全部或者部分内容、文字的真实性、完整性、及时性本站不做任何保证或承诺,请读者仅作参考,并请自行核实相关内容
版权声明:未经蚂蚁淘在线授权不得转载、摘编或利用其他方式使用上述作品。已经经本网授权使用作品的,应该授权范围内使用,并注明“来源:蚂蚁淘在线”。违反上述声明者,本网将追究其相关法律责任。





