- Higher yields of soluble proteins
- No interference with affinity purification resins
- Retain more protein activity
- Low viscosity without enzyme or sonication.
- Simple one-step protocol.
- Compatible with protease inhibitors, salts, chelating agents and reducing agents
The SoluLyse™ Protein Extraction Reagent is a proprietary formulation of nonionic detergents that is optimized for the most efficient extraction of soluble proteins from bacterial cells. The lysis reagent allows perforation of bacterial cell wall without denaturing proteins, and there is no need for secondary treatment such as sonication or freeze-thaw.
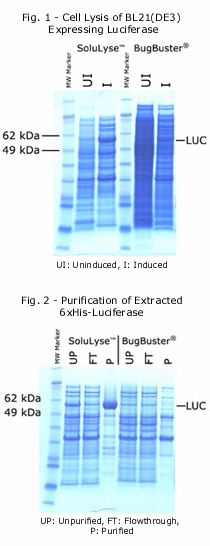
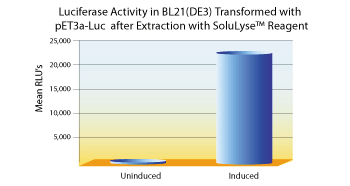
Side-by-side comparisons between SoluLyze andanother leading commercial lysis reagents showed that SoluLyse yields about 10-fold more soluble proteins. In addition, SoluLyse reagent does not compromise protein activities in cell lysates.
The SoluLyse reagent is easy to use. Simply add it to a bacterial pellet and gently mix for 10 minutes. The cells will be disrupted and soluble proteins released. Insoluble cell debris can then be removed by centrifugation. SoluLyse is also useful for the preparation of high-purity inclusion bodies when the protein of interest is insoluble.The SoluLyse Reagent is tested to be compatible with all popular purification methods, such as Ni and glutathione resins for purifying poly-His and GST-tagged proteins. The unique, mild formulation of SoluLyse offers minimized interference with the binding and elution processes and ensures superior purification results. In addition, the lysis reagent is compatible with a variety of additives such as protease inhibitors and reducing agents and may be readily dialyzed if needed.Contents- SoluLyse Protein Extraction Reagent is supplied in 1X solution in 50mM NaHPO4, pH 7.4.- SoluLyse Reagent in Tris Buffer is supplied in 1X solution in 20 mM Tris HCl, pH 7.5.
StorageStore at room temperature or +4ºC upon receipt. Stable for 2 years when stored properly.
New CitationsMuscadine grape skin extract inhibits prostate cancer cells by inducing cell-cycle arrest, and decreasing migration through heat shock protein 40.Ignacio, D. N.,et al (2019) Heliyon. 5(1): e01128.Link
Interplay of Klebsiella pneumoniae fabZ and lpxC Mutations Leads to LpxC Inhibitor-Dependent Growth Resulting from Loss of Membrane Homeostasis.Mostafavi, M.,et al (2018) mSphere. 3(5): e00508-18. Link
An open library of human kinase domain constructs for automated bacterial expression.Albanese, S. K.,et al (2018) Biochemistry. 57(31): 4675-4689. Link
Anacardic acid inhibits pancreatic cancer cell growth, and potentiates chemotherapeutic effect by Chmp1A -ATM - p53 signaling pathway.Park, M.,et al (2018) BMC Complement. Altern. Med. 18(1):71.Link
Multicenter Performance Assessment of Carba NP Test. Cunningham, S. A.,et al (2017) J. Clin. Microbiol. 55(6): 1954-1960. Link
Characterization of an Acinetobacter baumannii lptD Deletion Strain: Permeability Defects and Response to Inhibition of Lipopolysaccharide and Fatty Acid Biosynthesis.Bojkovic, J., et al (2016) J. Bacteriol. 198(4): 731-741. Link
Genomic mechanisms for cold tolerance and production of exopolysaccharides in the Arctic cyanobacterium Phormidesmis priestleyi BC1401.Chrismas, N. A. M.,et al (2016) BMC Genomics. 17: 533. Link
Toxic Accumulation of LPS Pathway Intermediates Underlies the Requirement of LpxH for Growth of Acinetobacter baumannii ATCC 19606.Richie, D. L.,et al (2016) PLoS. One. 11(8): e0160918. Link
RbsR Activates Capsule but Represses the rbsUDK Operon in Staphylococcus aureus.Lei, M. G.,et al (2015) J. Bacteriol. 197(23): 3666-3675. Link
Protein-Peptide Arrays for Detection of Specific Anti-Hepatitis D Virus (HDV) Genotype 1, 6, and 8 Antibodies among HDV-Infected Patients by Surface Plasmon Resonance Imaging.Villiers, M-B.,et al (2015) J. Clin. Microbiol. 53(4): 1164-71. Link
MicroRNA miR-182 cluster mediated modulation of RECK without changes in cell surface membrane type-1 matrix metalloproteinase (MT1-MMP).Silva, M.,et al (2015) Am. J. Cancer Res. 5(9): 2918-2928. Link