

- S Tag Peptide
- X-press Tag Peptide
- 3X FLAG Peptide
- Hexa His tag peptide
| FLAG tag PeptideVersatile fusion tag |

Sample solution is provided at 25 µL, 10mM.
- 1. Wang M, Hu J, et al. "High glucose-inducedubiquitination of G6PD leads to the injury of podocytes." FASEB J. 2019 Feb20:fj201801921R.PMID:30785802
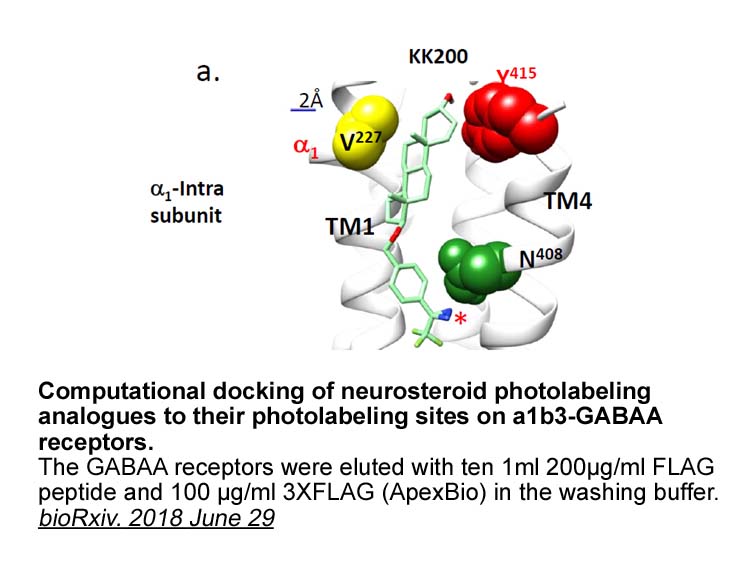
- 2. Zi-Wei Chen, John R Bracamontes, et al. "Multiple Functional Neurosteroid Binding Sites on GABAA Receptors." bioRxiv. 2018 June 29.
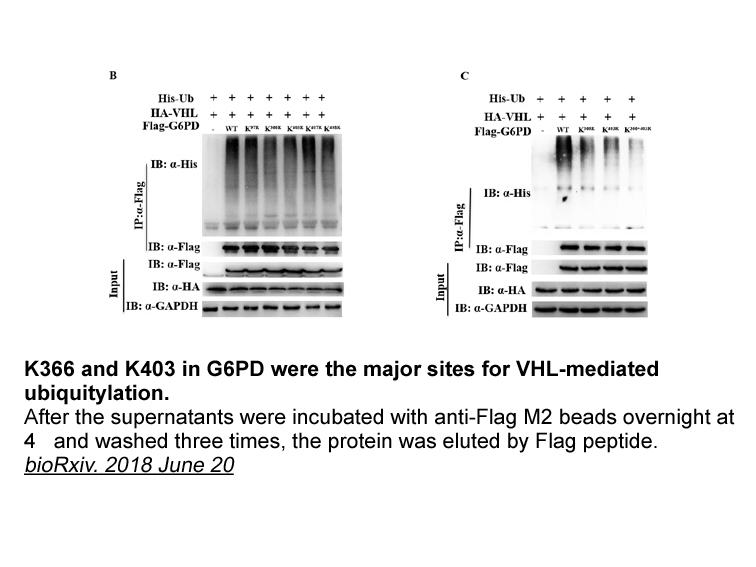
- 3. haoyun Zhang, Meng Wang, et al. "High glucose-induced ubiquitylation of G6PD leads to the injury of podocyte." bioRxiv. 2018 June 20.
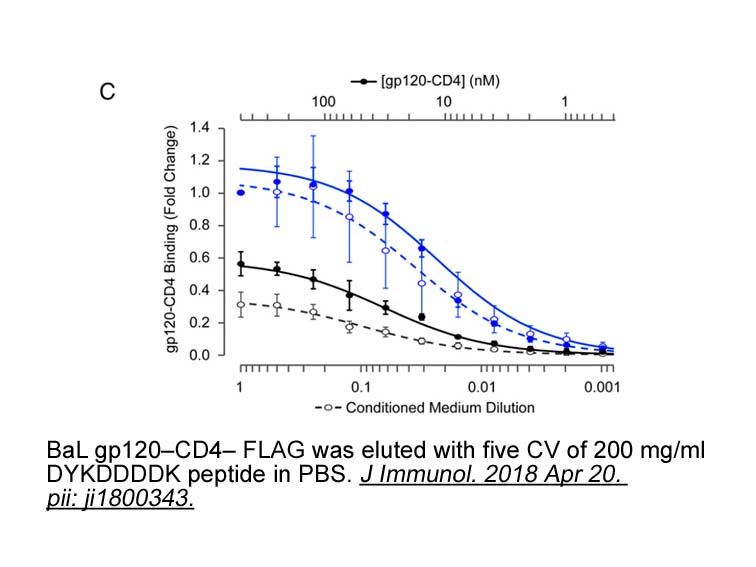
- 4. Heredia JD, Park J, et al. "Mapping Interaction Sites on Human Chemokine Receptors by Deep Mutational Scanning." J Immunol. 2018 Apr 20. pii: ji1800343.PMID:29678950
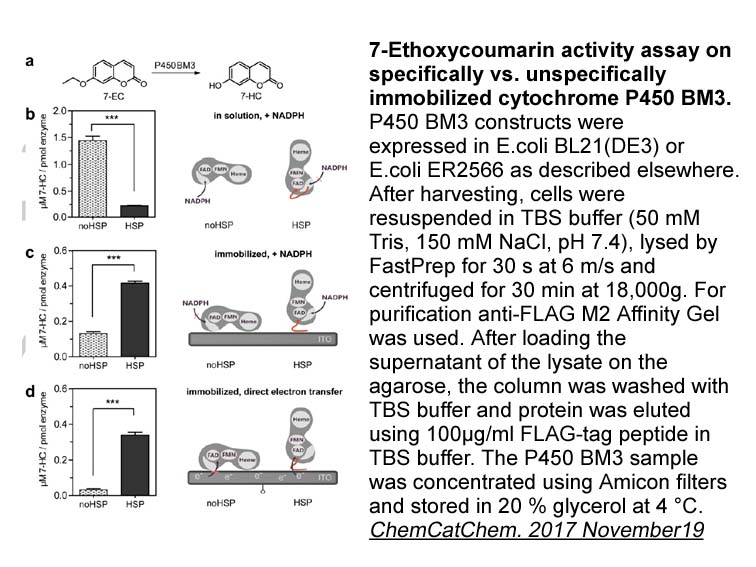
- 5. Sarah Zernia, Ronny Frank, et al. "Surface‐Binding Peptide Facilitates Electricity‐Driven NADPH‐Free Cytochrome P450 Catalysis." ChemCatChem. 2017 November19.
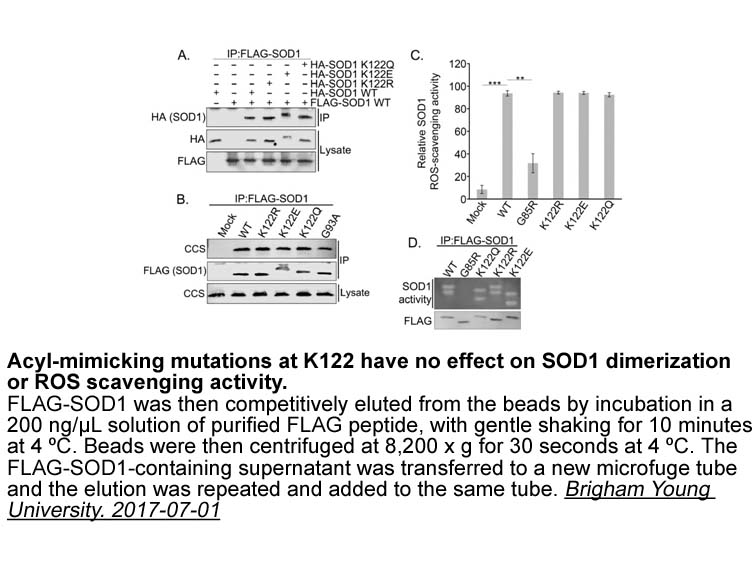
- 6. Banks CJ, Rodriguez NW, et al. "Acylation of Superoxide Dismutase 1 (SOD1) at K122 Governs SOD1-mediated Inhibition of Mitochondrial Respiration." Mol Cell Biol. 2017 Jul 24. pii: MCB.00354-17.PMID:28739857
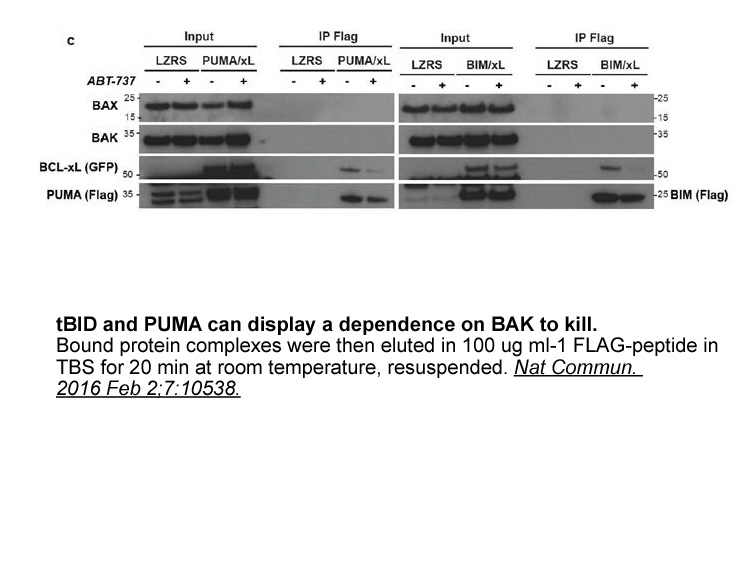
- 7. Lopez J, Bessou M, et al. "Mito-priming as a method to engineer Bcl-2 addiction." Nat Commun. 2016 Feb 2;7:10538.PMID:26833356
Quality Control & MSDS
- View current batch:
- Purity = 99.69%
- COA (Certificate Of Analysis)
- HPLC
- MS (Mass Spectrometry)
- MSDS (Material Safety Data Sheet)
- Datasheet
Chemical structure
Related Biological Data

Related Biological Data
Related Biological Data
Related Biological Data
Related Biological Data
Related Biological Data
Related Biological Data
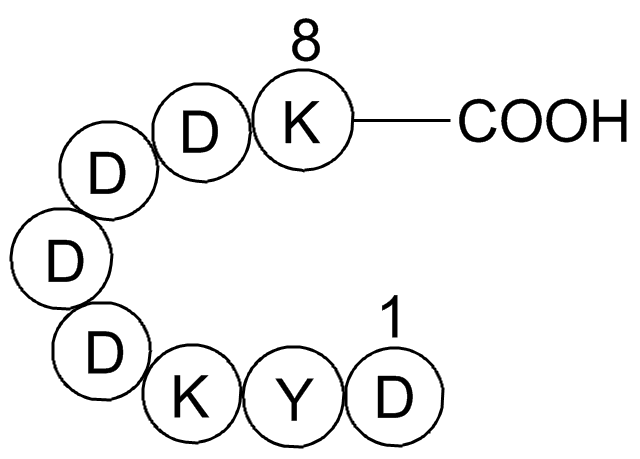
| Description | FLAG tag Peptide is a 8 amino acid peptide with an enterokinase-cleavage site used for the purification of recombinant proteins. | |||||
| Targets | anti-flag M2 antibody | |||||
| IC50 | ||||||

FLAG tag Peptide Dilution Calculator
calculate

FLAG tag Peptide Molarity Calculator
calculate
| Cas No. | 98849-88-8 | SDF | Download SDF |
| Synonyms | H-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-OH | ||
| Chemical Name | FLAG Peptide | ||
| Canonical SMILES | C1=CC(=CC=C1CC(C(=O)NC(CCCCN)C(=O)NC(CC(=O)O)C(=O)NC(CC(=O)O)C(=O)NC(CC(=O)O)C(=O)NC(CC(=O)O)C(=O)NC(CCCCN)C(=O)O)NC(=O)C(CC(=O)O)N)O | ||
| Formula | C41H60N10O20 | M.Wt | 1012.97 |
| Solubility | ≥50.6 mg/mL in DMSO, ≥210.6 mg/mL in H2O, ≥34.03 mg/mL in EtOH | Storage | Desiccate at -20°C |
| Physical Appearance | A solid | Shipping Condition | Evaluation sample solution : ship with blue ice.All other available size:ship with RT , or blue ice upon request |
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. | ||
A fusion tag called FLAG and consisting of eight amino acids Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys including an enterokinase-cleavage site, was specifically designed for immunoaffinity chromatography. It allows elution under non-denaturing conditions. Several antibodies against this peptide have been developed. One antibody, denoted as M1, binds the peptide in the presence of bivalent metal cations, preferably Ca+. Elution is effected by chelating agents. Another strategy is competitive elution with excess of free FLAG peptide. Antibodies M2 and M5 are applied in this procedure1.
The added marker segment is that it should not interfere with the native folding of proteins to which it is attached. Secondly, the marker peptide sequence should be water-soluble and should retain a high degree of exposure on the surface of the protein, so that it can readily interact with its ligand. It should also be suitable for a mild and inexpensive affinity purification procedure. Finally, an easy removal of the marker peptide leading to a native product is also advantageous. Due to this small size, the marker peptide can be encoded by a single synthetic oligonucleotide. It is known that aromatic amino acids are the major factors in antigen–antibody interactions.
Lys at position 3 in the marker sequence leads to a hexapeptide sequence LysAspAspAspAspLys, which ensures a maximum value on the hydrophilicity scale according to Hopp and Woods2 . Such hydrophilic sequences have been shown to express strong antigenicity and are thus likely to adopt a highly exposed conformation in the three-dimensional folding of proteins3. Another virtue of FLAG is that the longest trypsinogen prosequences are of this length. This enables the removal of the tag and the production of an authentic N-terminus of the fusion protein partner by enterokinase treatment. The FLAG peptide can be fused to either the N- or C-terminus of a given fusion protein. Nevertheless, the N-terminal fusion has several advantages. Inhibition ELISA experiments showed that the anti-Flag antibody M1 binds three to four orders of magnitude better under conditions where the a-amino group of the first amino acid is freely accessible4.
The FLAG marker peptide fusion system comprises a unique and widely useful technique for protein identification and purification. Elution of the fusion protein can be accomplished either by antibody-mediated affinity chromatography in a calcium-dependent manner, by lowering the pH, or by competitive elution with synthetic peptides1. Although highly selective, the binding capacities are low, making scale-up a costly undertaking. In addition to cost and low capacity, large-scale immunoaffinity chromatography, applied to the production of therapeutic proteins has several disadvantages: ligand leakage, instability, and need for validation of antibody production. The stability of the affinity chromatography column depends on the nature and source of the crude extracts. Furthermore, the FLAGe tag, designed to be immunogenic, must be removed from therapeutic proteins. In most cases, this can be accomplished with enterokinase. However, contaminating proteases may also produce undesired cleavages. Despite these drawbacks, the FLAG fusion is useful in research and development: FLAG proteins can be readily purified and assayed by ELISA or any other immunochemical detection method, thus expediting the raising of antisera against a desired protein and characterization studies.
References:1. A. Einhauer, A. Jungbauer. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49 2001 455–465. 2. Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A 1981;78:3824–8.3. Hopp TP. Protein surface analysis: methods for identifying antigenic determinants and other interaction sites. J Immunol Methods 1986;88:1–18.4. Power BE, Ivancic N, Harley VR, Webster RG, Kortt AA, Irving RA, et al. High-level temperature-induced synthesis of an a




