

 )
)Asp-Ile-Ser-Gly-Ile-Asn-Ala-Ser-Val-Val-Asn-Ile-Gln-Lys-Glu-Ile-Asp-Arg-Leu-Asn-Glu-Val-Ala-Lys-Asn-Leu-Asn-Glu-Ser-Leu-Ile-Asp-Leu-Gln-Glu-Leu |
- Molecular Weight:
- Purity:
- Validation:
- Storage:
- Appearance:
- Contents:
| 4008.50 | |
| ≥ 95% | |
| Exhibits correct molecular weight | |
| Up to 6 months in lyophilized form at 0-5ºC. For best results, rehydrate just before use. After rehydration, freeze at -20ºC for up to 3 months. Aliquot before freezing to avoid repeated freeze-thaw cycles. | |
| White powder | |
| Each vial contains 100 μg of NET peptide. |
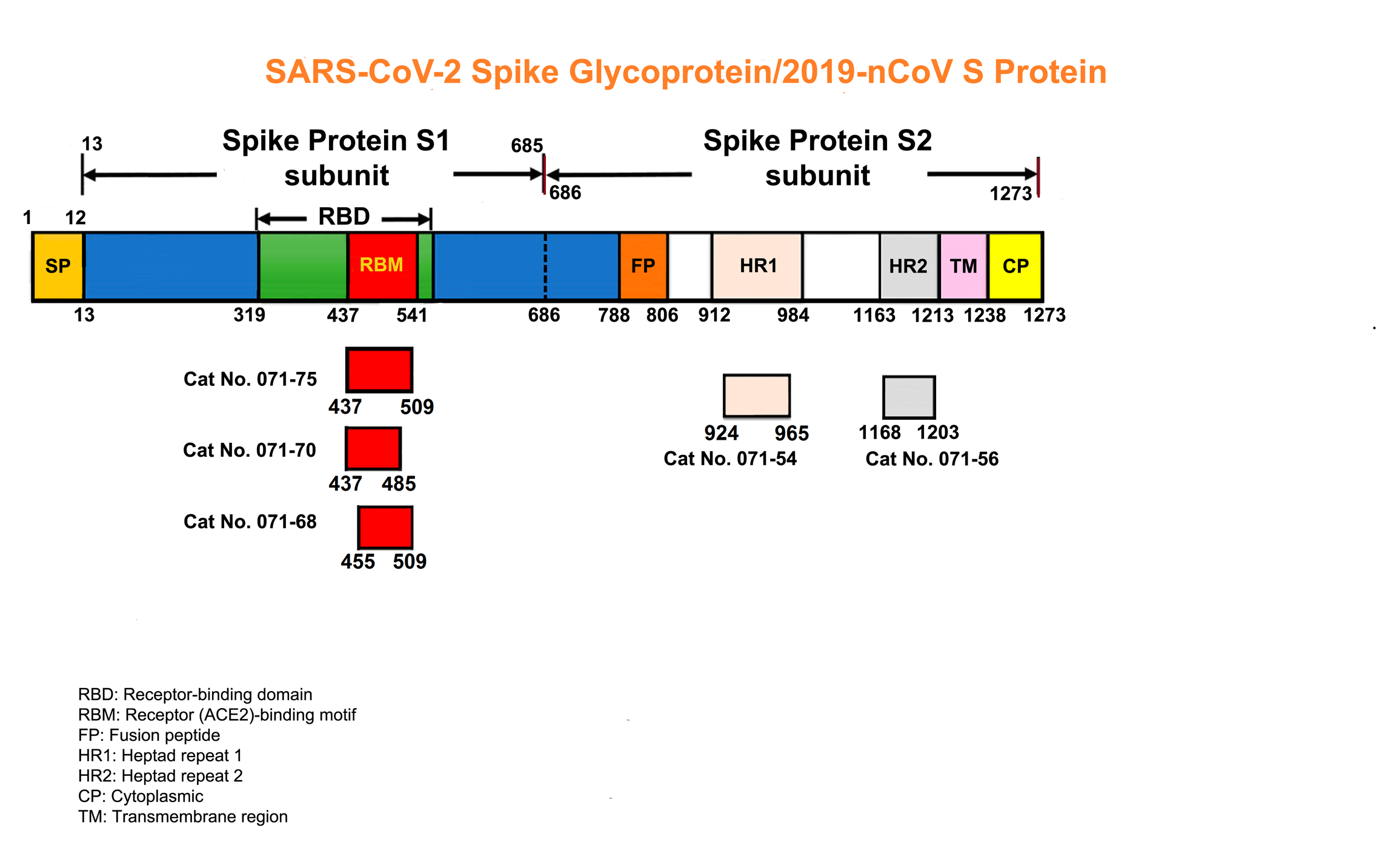
RBD of SARS-COV-2 S Protein, EK1 and ACE2 enzyme receptor peptides help to understand COVID-19
- Abstracts
- Schematics
- More Information


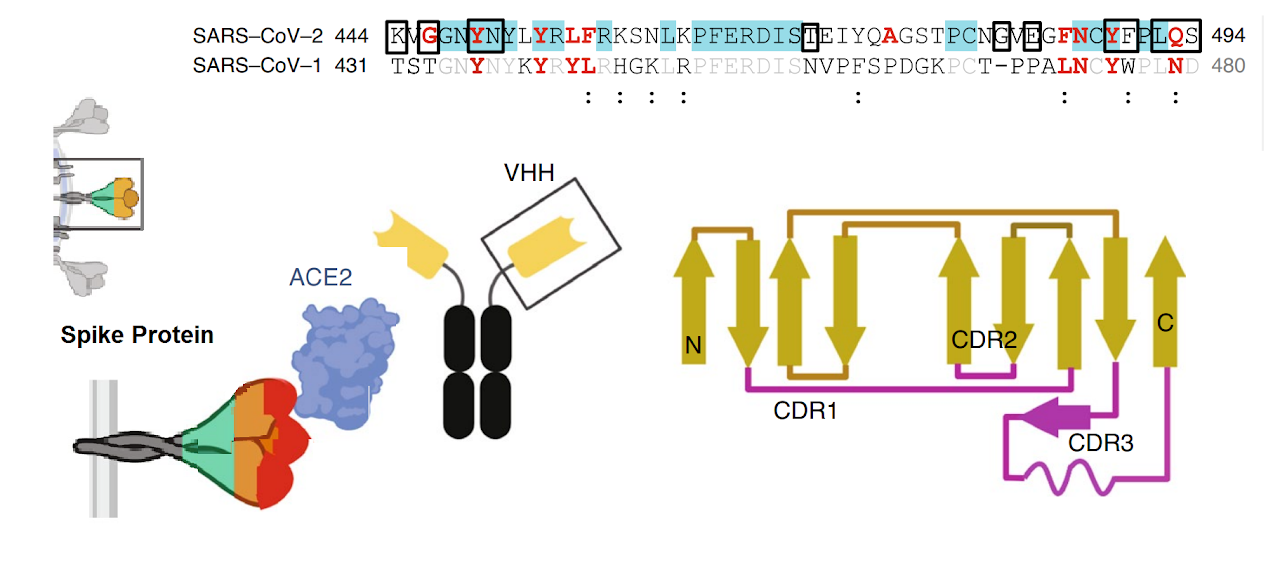
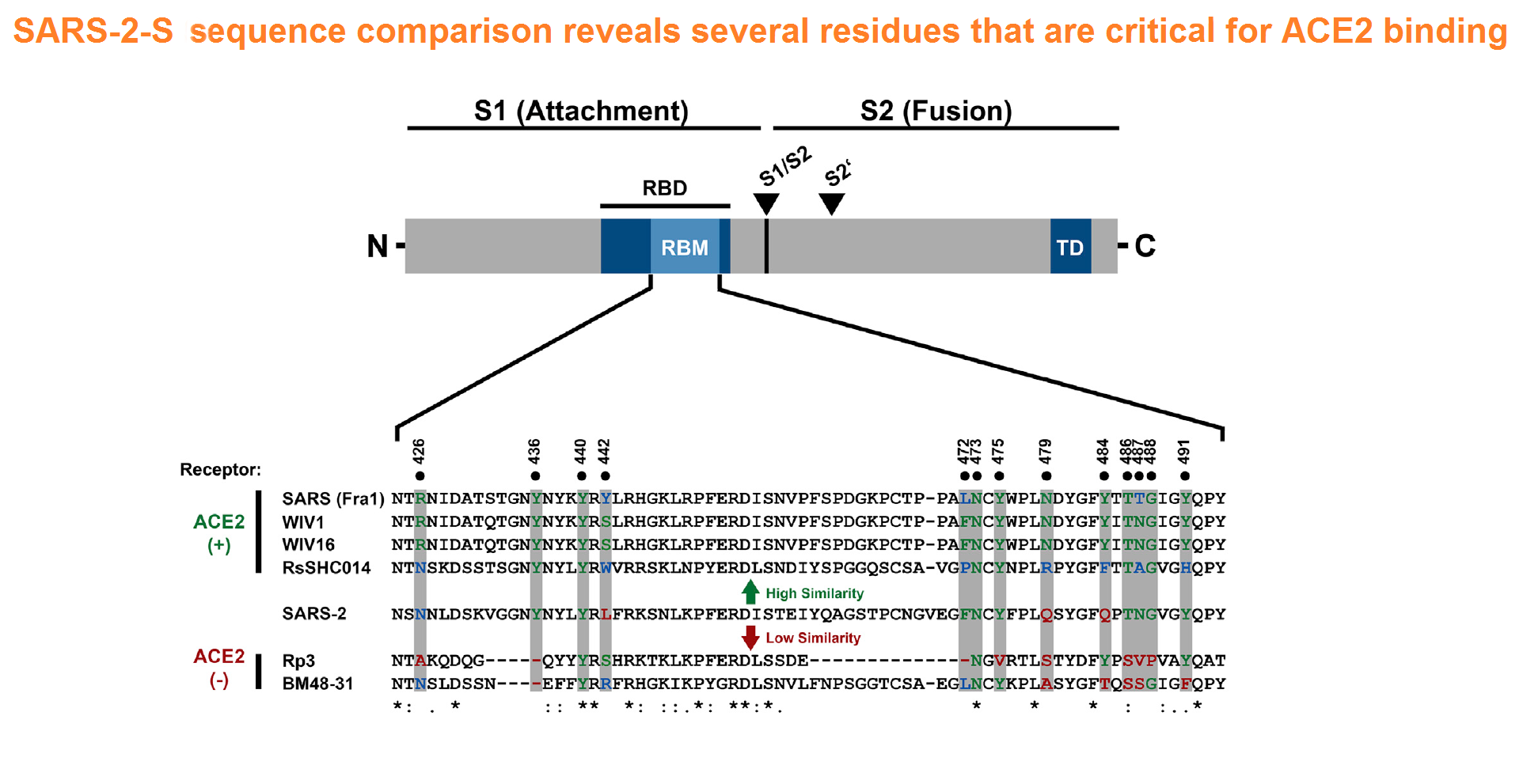
The spike protein of SARS-CoV-2 drives infection. The spike protein is composed of S1 and S2 subunits. S1 contains the RBD (highlighted in red). Using the RBD, the trimeric spoke molecule binds to ACE2 on human cells (a single ACE2 is shown in blue). RBD residues that are shared between SARS-CoV-2 and SARS-Cov-1 are highlited in cyan (SARS-CoV-2 top) or in light gray text (SARS-CoV-1, bottom). Resideus that contact ACE2 are highlited in red for each sequence. Other sequence differences are in black test, with conservative substitutions indivated by colons (:) underneath. Resideies in contact with the H11-H4 nanobody are boxed.
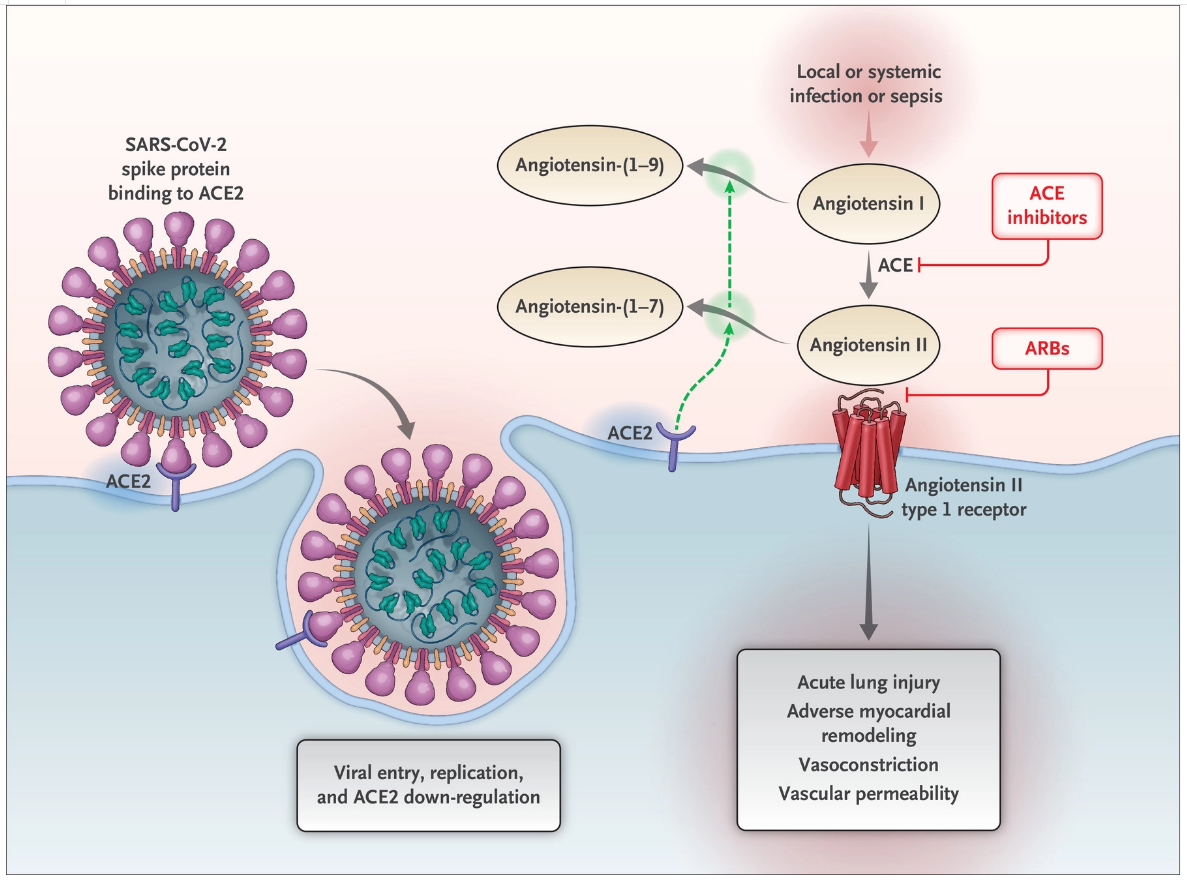
Figure from Jiangdong Huo et al. ofNat Struct Mol Biol . 2020 Jul 13. doi: 10.1038/s41594-020-0469-6A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm
Abstract: Neither the disease mechanism nor treatments for COVID-19 are currently known. Here, we present a novel molecular mechanism for COVID-19 that provides therapeutic intervention points that can be addressed with existing FDA-approved pharmaceuticals. The entry point for the virus is ACE2, which is a component of the counteracting hypotensive axis of RAS. Bradykinin is a potent part of the vasopressor system that induces hypotension and vasodilation and is degraded by ACE and enhanced by the angiotensin1-9produced by ACE2. Here, we perform a new analysis on gene expression data from cells in bronchoalveolar lavage fluid (BALF) from COVID-19 patients that were used to sequence the virus. Comparison with BALF from controls identifies a critical imbalance in RAS represented by decreased expression of ACE in combination with increases in ACE2, renin, angiotensin, key RAS receptors, kinogen and many kallikrein enzymes that activate it, and both bradykinin receptors. This very atypical pattern of the RAS is predicted to elevate bradykinin levels in multiple tissues and systems that will likely cause increases in vascular dilation, vascular permeability and hypotension. These bradykinin-driven outcomes explain many of the symptoms being observed in COVID-19.Garvin MR, Alvarez C, Miller JI, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;9
Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome
Abstract: COVID-19 patients can present with pulmonary edema early in disease. We propose that this is due to a local vascular problem because of activation of bradykinin 1 receptor (B1R) and B2R on endothelial cells in the lungs. SARS-CoV-2 enters the cell via ACE2 that next to its role in RAAS is needed to inactivate des-Arg9 bradykinin, the potent ligand of the B1R. Without ACE2 acting as a guardian to inactivate the ligands of B1R, the lung environment is prone for local vascular leakage leading to angioedema. Here, we hypothesize that a kinin-dependent local lung angioedema via B1R and eventually B2R is an important feature of COVID-19. We propose that blocking the B2R and inhibiting plasma kallikrein activity might have an ameliorating effect on early disease caused by COVID-19 and might prevent acute respiratory distress syndrome (ARDS). In addition, this pathway might indirectly be responsive to anti-inflammatory agents.Van de veerdonk FL, Netea MG, Van deuren M, et al. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife. 2020;9
Outcomes Associated With Use of a Kinin B2 Receptor Antagonist Among Patients With COVID-19
Abstract: This case-control study examines the association between receipt of the bradykinin 2 (B2) receptor antagonist icatibant and improved oxygenation in patients with coronavirus disease 2019 (COVID-19).Van de veerdonk FL, Kouijzer IJE, De nooijer AH, et al. Outcomes Associated With Use of a Kinin B2 Receptor Antagonist Among Patients With COVID-19. JAMA Netw Open. 2020;3(8):e2017708.
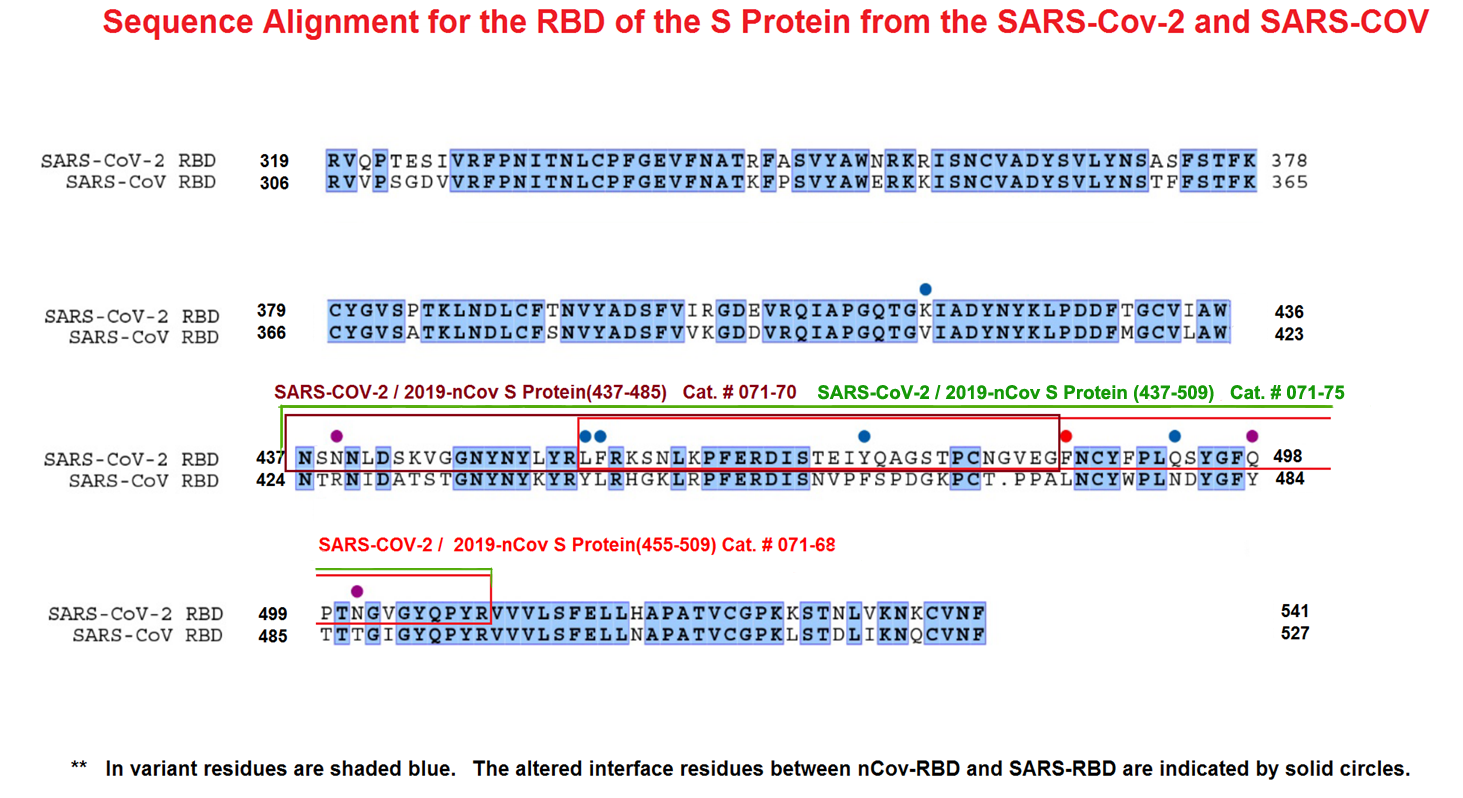
Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine
Abstract: The outbreak of Coronavirus Disease 2019 (COVID-19) has posed a serious threat to global public health, calling for the development of safe and effective prophylactics and therapeutics against infection of its causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as 2019 novel coronavirus (2019-nCoV). The CoV spike (S) protein plays the most important roles in viral attachment, fusion and entry, and serves as a target for development of antibodies, entry inhibitors and vaccines. Here, we identified the receptor-binding domain (RBD) in SARS-CoV-2 S protein and found that the RBD protein bound strongly to human and bat angiotensin-converting enzyme 2 (ACE2) receptors. SARS-CoV-2 RBD exhibited significantly higher binding affinity to ACE2 receptor than SARS-CoV RBD and could block the binding and, hence, attachment of SARS-CoV-2 RBD and SARS-CoV RBD to ACE2-expressing cells, thus inhibiting their infection to host cells. SARS-CoV RBD-specific antibodies could cross-react with SARS-CoV-2 RBD protein, and SARS-CoV RBD-induced antisera could cross-neutralize SARS-CoV-2, suggesting the potential to develop SARS-CoV RBD-based vaccines for prevention of SARS-CoV-2 and SARS-CoV infection.Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020
Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2
Abstract: The recent emergence of a novel coronavirus (SARS-CoV-2) in China has caused significant public health concerns. Recently, ACE2 was reported as an entry receptor for SARS-CoV-2. In this study, we present the crystal structure of the C-terminal domain of SARS-CoV-2 (SARS-CoV-2-CTD) spike (S) protein in complex with human ACE2 (hACE2), which reveals a hACE2-binding mode similar overall to that observed for SARS-CoV. However, atomic details at the binding interface demonstrate that key residue substitutions in SARS-CoV-2-CTD slightly strengthen the interaction and lead to higher affinity for receptor binding than SARS-RBD. Additionally, a panel of murine monoclonal antibodies (mAbs) and polyclonal antibodies (pAbs) against SARS-CoV-S1/receptor-binding domain (RBD) were unable to interact with the SARS-CoV-2 S protein, indicating notable differences in antigenicity between SARS-CoV and SARS-CoV-2. These findings shed light on the viral pathogenesis and provide important structural information regarding development of therapeutic countermeasures against the emerging virus.Wang Q, Zhang Y, Wu L, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020
Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV
Abstract: Since 2002, beta coronaviruses (CoV) have caused three zoonotic outbreaks, SARS-CoV in 2002–2003, MERS-CoV in 2012, and the newly emerged SARS-CoV-2 in late 2019. However, little is currently known about the biology of SARS-CoV-2. Here, using SARS-CoV-2 S protein pseudovirus system, we confirm that human angiotensin converting enzyme 2 (hACE2) is the receptor for SARS-CoV-2, find that SARS-CoV-2 enters 293/hACE2 cells mainly through endocytosis, that PIKfyve, TPC2, and cathepsin L are critical for entry, and that SARS-CoV-2 S protein is less stable than SARS-CoV S. Polyclonal anti-SARS S1 antibodies T62 inhibit entry of SARS-CoV S but not SARS-CoV-2 S pseudovirions. Further studies using recovered SARS and COVID-19 patients’ sera show limited cross-neutralization, suggesting that recovery from one infection might not protect against the other. Our results present potential targets for development of drugs and vaccines for SARS-CoV-2.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
Abstract: The recent emergence of the novel, pathogenic SARS-coronavirus 2 (SARS-CoV-2) in China and its rapid national and international spread pose a global health emergency. Cell entry of coronaviruses depends on binding of the viral spike (S) proteins to cellular receptors and on S protein priming by host cell proteases. Unravelling which cellular factors are used by SARS-CoV-2 for entry might provide insights into viral transmission and reveal therapeutic targets. Here, we demonstrate that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming. A TMPRSS2 inhibitor approved for clinical use blocked entry and might constitute a treatment option. Finally, we show that the sera from convalescent SARS patients cross-neutralized SARS-2-S-driven entry. Our results reveal important commonalities between SARS-CoV-2 and SARS-CoV infection and identify a potential target for antiviral intervention.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271-280.e8.
Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor
Abstract: A new and highly pathogenic coronavirus (severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)) caused an outbreak in Wuhan city, Hubei province, China starting from December 2019 that quickly spread nationwide and to other countries around the world1-3. Here, to better understand the initial step of infection at an atomic level, we determined the crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 bound to the cell receptor ACE2. The overall ACE2-binding mode of the SARS-CoV-2 RBD is nearly identical to that of the SARS-CoV RBD, which also uses ACE2 as the cell receptor4. Structural analysis identified residues in the SARS-CoV-2 RBD that are essential for ACE2 binding, the majority of which either are highly conserved or share similar side chain properties with those in the SARS-CoV RBD. Such similarity in structure and sequence strongly indicate convergent evolution between the SARS-CoV-2 and SARS-CoV RBDs for improved binding to ACE2, although SARS-CoV-2 does not cluster within SARS and SARS-related coronaviruses1-3,5. The epitopes of two SARS-CoV antibodies that target the RBD are also analysed for binding to the SARS-CoV-2 RBD, providing insights into the future identification of cross-reactive antibodies.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;
Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2
Abstract: Angiotensin-converting enzyme 2 (ACE2) is the cellular receptor for severe acute respiratory syndrome-coronavirus (SARS-CoV) and the new coronavirus (SARS-CoV-2) that is causing the serious coronavirus disease 2019 (COVID-19) epidemic. Here, we present cryo-electron microscopy structures of full-length human ACE2 in the presence of the neutral amino acid transporter B0AT1 with or without the receptor binding domain (RBD) of the surface spike glycoprotein (S protein) of SARS-CoV-2, both at an overall resolution of 2.9 angstroms, with a local resolution of 3.5 angstroms at the ACE2-RBD interface. The ACE2-B0AT1 complex is assembled as a dimer of heterodimers, with the collectrin-like domain of ACE2 mediating homodimerization. The RBD is recognized by the extracellular peptidase domain of ACE2 mainly through polar residues. These findings provide important insights into the molecular basis for coronavirus recognition and infection.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444-8.
Renin–Angiotensin–Aldosterone System Inhibitors in Patients with Covid-19
The renin–angiotensin–aldosterone system (RAAS) is an elegant cascade of vasoactive peptides that orchestrate key processes in human physiology. Severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) and SARS-CoV-2, which have been responsible for the SARS epidemic in 2002 to 2004 and for the more recent coronavirus disease 2019 (Covid-19) pandemic, respectively, interface with the RAAS through angiotensin-converting enzyme 2 (ACE2), an enzyme that physiologically counters RAAS activation but also functions as a receptor for both SARS viruses.1,2 The interaction between the SARS viruses and ACE2 has been proposed as a potential factor in their infectivity,3,4 and there are concerns about the use of RAAS inhibitors that may alter ACE2 and whether variation in ACE2 expression may be in part responsible for disease virulence in the ongoing Covid-19 pandemic.5-8 Indeed, some media sources and health systems have recently called for the discontinuation of ACE inhibitors and angiotensin-receptor blockers (ARBs), both prophylactically and in the context of suspected Covid-19.Vaduganathan, M., Vardeny, O., Michel, T., McMurray, J. J. V., Pfeffer, M. A., & Solomon, S. D. (2020). Renin–Angiotensin–Aldosterone System Inhibitors in Patients with Covid-19. New England Journal of Medicine.
Organ-protective Effect of Angiotensin-Converting Enzyme 2 and Its Effect on the Prognosis of COVID-19
Abstract: This article reviews the correlation between angiotensin-converting enzyme 2 (ACE2) and severe risk factors for coronavirus disease 2019 (COVID-19) and the possible mechanisms. ACE2 is a crucial component of the renin-angiotensin system (RAS). The classical RAS ACE-Ang II-AT1R regulatory axis and the ACE2-Ang 1-7-MasR counter-regulatory axis play an essential role in maintaining homeostasis in humans. ACE2 is widely distributed in the heart, kidneys, lungs, and testes. ACE2 antagonizes the activation of the classical RAS system and protects against organ damage, protecting against hypertension, diabetes, and cardiovascular disease. Similar to SARS-CoV, SARS-CoV-2 also uses the ACE2 receptor to invade human alveolar epithelial cells. Acute respiratory distress syndrome (ARDS) is a clinical high-mortality disease, and ACE2 has a protective effect on this type of acute lung injury. Current research shows that the poor prognosis of patients with COVID-19 is related to factors such as sex (male), age (>60 years), underlying diseases (hypertension, diabetes, and cardiovascular disease), secondary ARDS, and other relevant factors. Because of these protective effects of ACE2 on chronic underlying diseases and ARDS, the development of spike protein-based vaccine and drugs enhancing ACE2 activity may become one of the most promising approaches for the treatment of COVID-19 in the future.Cheng, H., Wang, Y., & Wang, G.-Q. (2020). Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. Journal of Medical Virology.
Inhibition of SARS-CoV-2 infections in engineered human cell byclinical grade soluble ACE2
Summary: We have previously provided the first genetic evidence that Angiotensin converting enzyme 2 (ACE2) is the critical receptor for SARS-CoV and that ACE2 protects the lung from injury, providing a molecular explanation for the severe lung failure and death due to SARS-CoV infections. ACE2 has now also been identified as a key receptor for SARS-CoV-2 infections and it has been proposed that inhibiting this interaction might be used in treating patients with COVID19. However, it is not known whether human recombinant soluble ACE2 (hrsACE2) blocks growth of SARS-CoV-2. Here we show that clinical grade hrsACE2 reduced SARS-CoV-2 recovery from Vero cells by a factor of 1,000-5,000. An equivalent mouse rsACE2 had no effect. We also show that SARS-CoV-2 can directly infect engineered human blood vessel organoids and human kidney organoids, which can be inhibited by hrsACE2. These data demonstrate that hrsACE2 can significantly block early stages of SARS-CoV-2 infections.Monteil et al, Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2, Cell 2020, DOI: 10.1016/j.cell.2020.04.004
The first-in-class peptide binder to the SARS-CoV-2 spike protein
Abstract: Coronavirus disease 19 (COVID-19) is an emerging global health crisis. With over 200,000 confirmed cases to date, this pandemic continues to expand, spurring research to discover vaccines and therapies. SARS-CoV-2 is the novel coronavirus responsible for this disease. It initiates entry into human cells by binding to angiotensin-converting enzyme 2 (ACE2) via the receptor binding domain (RBD) of its spike protein (S). Disrupting the SARS-CoV-2-RBD binding to ACE2 with designer drugs has the potential to inhibit the virus from entering human cells, presenting a new modality for therapeutic intervention. Peptide-based binders are an attractive solution to inhibit the RBD-ACE2 interaction by adequately covering the extended protein contact interface. Using molecular dynamics simulations based on the recently solved ACE2 and SARS CoV-2-RBD co-crystal structure, we observed that the ACE2 peptidase domain (PD) α1 helix is important for binding SARS-CoV-2-RBD. Using automated fast-flow peptide synthesis, wechemically synthesized a 23-mer peptide fragment of the ACE2 PD α1 helix composed entirely ofproteinogenic amino acids. Chemical synthesis of this human derived sequence was complete in1.5 hours and after work up and isolation >20 milligrams of pure material was obtained. Bio-layer interferometry revealed that this peptide specifically associates with the SARS-CoV-2-RBD with low nanomolar affinity. This peptide binder to SARS-CoV-2-RBD provides new avenues forCOVID-19 treatment and diagnostic modalities by blocking the SARS-CoV-2 spike proteininteraction with ACE2 and thus precluding virus entry into human cells.Zhang, G., Pomplun, S., Loftis, A. R., Loas, A., & Pentelute, B. L. (2020). The first-in-class peptide binder to the SARS-CoV-2 spike protein. Cold Spring Harbor Laboratory.
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Abstract: The recent outbreak of coronavirus disease (COVID-19) caused by SARS-CoV-2 infection in Wuhan, China has posed a serious threat to global public health. To develop specific anti-coronavirus therapeutics and prophylactics, the molecular mechanism that underlies viral infection must first be defined. Therefore, we herein established a SARS-CoV-2 spike (S) protein-mediated cell–cell fusion assay and found that SARS-CoV-2 showed a superior plasma membrane fusion capacity compared to that of SARS-CoV. We solved the X-ray crystal structure of six-helical bundle (6-HB) core of the HR1 and HR2 domains in the SARS-CoV-2 S protein S2 subunit, revealing that several mutated amino acid residues in the HR1 domain may be associated with enhanced interactions with the HR2 domain. We previously developed a pan-coronavirus fusion inhibitor, EK1, which targeted the HR1 domain and could inhibit infection by divergent human coronaviruses tested, including SARS-CoV and MERS-CoV. Here we generated a series of lipopeptides derived from EK1 and found that EK1C4 was the most potent fusion inhibitor against SARS-CoV-2 S protein-mediated membrane fusion and pseudovirus infection with IC50s of 1.3 and 15.8 nM, about 241- and 149-fold more potent than the original EK1 peptide, respectively. EK1C4 was also highly effective against membrane fusion and infection of other human coronavirus pseudoviruses tested, including SARS-CoV and MERS-CoV, as well as SARSr-CoVs, and potently inhibited the replication of 5 live human coronaviruses examined, including SARS-CoV-2. Intranasal application of EK1C4 before or after challenge with HCoV-OC43 protected mice from infection, suggesting that EK1C4 could be used for prevention and treatment of infection by the currently circulating SARS-CoV-2 and other emerging SARSr-CoVs.Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV)infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343-355.
Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies
Abstract: The beginning of 2020 has seen the emergence of COVID-19 outbreak caused by a novel coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). There is an imminent need to better understand this new virus and to develop ways to control its spread. In this study, we sought to gain insights for vaccine design against SARS-CoV-2 by considering the high genetic similarity between SARS-CoV-2 and SARS-CoV, which caused the outbreak in 2003, and leveraging existing immunological studies of SARS-CoV. By screening the experimentally-determined SARS-CoV-derived B cell and T cell epitopes in the immunogenic structural proteins of SARS-CoV, we identified a set of B cell and T cell epitopes derived from the spike (S) and nucleocapsid (N) proteins that map identically to SARS-CoV-2 proteins. As no mutation has been observed in these identified epitopes among the 120 available SARS-CoV-2 sequences (as of 21 February 2020), immune targeting of these epitopes may potentially offer protection against this novel virus. For the T cell epitopes, we performed a population coverage analysis of the associated MHC alleles and proposed a set of epitopes that is estimated to provide broad coverage globally, as well as in China. Our findings provide a screened set of epitopes that can help guide experimental efforts towards the development of vaccines against SARS-CoV-2.Ahmed SF, Quadeer AA, Mckay MR. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12(3)
The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade
Abstract: In 2019, a new coronavirus (2019-nCoV) infecting Humans has emerged in Wuhan, China. Its genome has been sequenced and the genomic information promptly released. Despite a high similarity with the genome sequence of SARS-CoV and SARS-like CoVs, we identified a peculiar furin-like cleavage site in the Spike protein of the 2019-nCoV, lacking in the other SARS-like CoVs. In this article, we discuss the possible functional consequences of this cleavage site in the viral cycle, pathogenicity and its potential implication in the development of antivirals.Coutard B, Valle C, De lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742.
Drug Treatment Options for the 2019-new Coronavirus (2019-nCoV)
Abstract: As of January 22, 2020, a total of 571 cases of the 2019-new coronavirus (2019-nCoV) have been reported in 25 provinces (districts and cities) in China. At present, there is no vaccine or antiviral treatment for human and animal coronavirus, so that identifying the drug treatment options as soon as possible is critical for the response to the 2019-nCoV outbreak. Three general methods, which include existing broad-spectrum antiviral drugs using standard assays, screening of a chemical library containing many existing compounds or databases, and the redevelopment of new specific drugs based on the genome and biophysical understanding of individual coronaviruses, are used to discover the potential antiviral treatment of human pathogen coronavirus. Lopinavir /Ritonavir, Nucleoside analogues, Neuraminidase inhibitors, Remdesivir, peptide (EK1), arbidol, RNA synthesis inhibitors (such as TDF, 3TC), anti-inflammatory drugs (such as hormones and other molecules), Chinese traditional medicine, such ShuFengJieDu Capsules and Lianhuaqingwen Capsule, could be the drug treatment options for 2019-nCoV. However, the efficacy and safety of these drugs for 2019- nCoV still need to be further confirmed by clinical experiments.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends. 2020;
Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein
Introduction:Very recently, a novel coronavirus, 2019-nCoV, emerged in Wuhan, China and then quickly spread worldwide, resulting in >17,388 confirmed cases and 361 deaths as of 3 February 2020, thus calling for the development of safe and effective therapeutics and prophylatics.Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;
A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike
Abstract: Continuously emerging highly pathogenic human coronaviruses (HCoVs) remain a major threat to human health, as illustrated in past SARS-CoV and MERS-CoV outbreaks. The development of a drug with broad-spectrum HCoV inhibitory activity would address this urgent unmet medical need. Although previous studies have suggested that the HR1 of HCoV spike (S) protein is an important target site for inhibition against specific HCoVs, whether this conserved region could serve as a target for the development of broad-spectrum pan-CoV inhibitor remains controversial. Here, we found that peptide OC43-HR2P, derived from the HR2 domain of HCoV-OC43, exhibited broad fusion inhibitory activity against multiple HCoVs. EK1, the optimized form of OC43-HR2P, showed substantially improved pan-CoV fusion inhibitory activity and pharmaceutical properties. Crystal structures indicated that EK1 can form a stable six-helix bundle structure with both short α-HCoV and long β-HCoV HR1s, further supporting the role of HR1 region as a viable pan-CoV target site.Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580.
Identification of Critical Determinants on ACE2 for SARS-CoV Entry and Development of a Potent Entry Inhibitor.
Abstract: Severe acute respiratory syndrome (SARS) is caused by a novel coronavirus, SARS-CoV. Virus entry into cells is mediated through interactions between spike (S) glycoprotein and angiotensin-converting enzyme 2 (ACE2). Alanine scanning mutagenesis analysis was performed to identify determinants on ACE2 critical for SARS-CoV infection. Results indicated that charged amino acids between residues 22 and 57 were important, K26 and D30, in particular. Peptides representing various regions of ACE2 critical for virus infection were chemically synthesized and evaluated for antiviral activity. Two peptides (a.a. 22-44 and 22-57) exhibited a modest antiviral activity with IC50 of about 50 microM and 6 microM, respectively. One peptide comprised of two discontinuous segments of ACE2 (a.a. 22-44 and 351-357) artificially linked together by glycine, exhibited a potent antiviral activity with IC50 of about 0.1 microM. This novel peptide is a promising candidate as a therapeutic agent against this deadly emerging pathogen.Han, D. P., Penn-Nicholson, A., & Cho, M. W. (2006). Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology, 350(1), 15–25
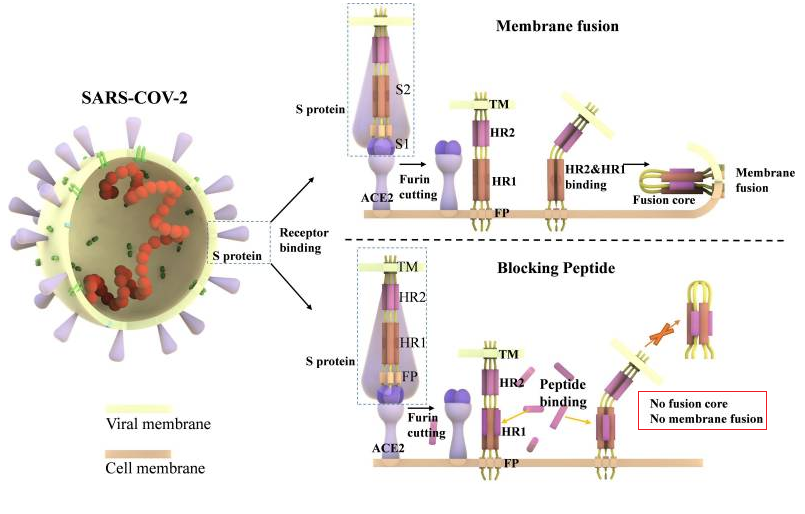
Figure 1. Ling R, Dai Y, Huang B, Huang W, Lu X, Jiang Y. In Silico Design of Antiviral Peptides Targeting the Spike Protein of SARS-CoV-2. Peptides. 2020;:170328.
No References




